
Evolution and regulation of natural product biosynthesis
In our group we aim to (i) better understand how plants repeatedly evolve ways to produce the same type of natural products and (ii) to apply and adapt new methods to study pathways and regulation at the cell type specific level in model and non-model species.

Plants produce an enormous diversity of natural products (specialized metabolites). While some of them are unique and only occur in specific lineages others are more widespread. Interestingly, in some cases the biosynthesis of even complex natural products was invented more than once in nature. Studying such convergently evolved metabolic pathways offers not only unique insights into pathway evolution but can also inform pathway engineering approaches.
A remarkable example of a convergently evolved complex natural product scaffold are ipecac alkaloids. These monoterpenoid-derived tetrahydroisoquinoline alkaloids occur in the Cornales and Gentianales orders which diverged at least 100 million years ago. In our group we study ipecac alkaloid biosynthesis in Carapichea ipecauanha (Gentianales) and Alangium salviifolium (Cornales) in a comparative manner as a model system for convergent pathway evolution. Noteworthy, some ipecac alkaloids have emetic activity and therefore both species are well known medicinal plants: “Ipecac syrup”, an extract prepared from C. ipecacuanha roots, was an over-the-counter emetic drug used for many decades, while A. salviifolium is known as “Ankol” in traditional ayurvedic medicine to treat intestinal parasite infections, rheumatism and many other diseases.
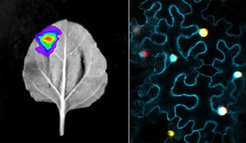
We have been elucidating the pathways in both species using classic pathway discovery methods i.e. generating genomics, transcriptomics and metabolomics data and subsequent pathway reconstitution in Nicotiana benthamiana as well as in vitro enzymatic activity assays using recombinantly produced proteins. In silico approaches such as phylogenetic analyses and protein model comparisons (alphafold2) are used to compare and contrast the pathway enzymes and to unravel from which primary metabolism enzyme families they were recruited. Additionally, we tackle the important but currently less commonly studied aspect of evolution of regulatory mechanisms of pathways. For instance, pathway gene expression, as well as enzyme subcellular localization and potential metabolic channeling must be coordinated to achieve an efficient production of a natural product. Hence, we also compare and contrast gene expression pattern from the tissue down to the single cell level in both species and analyze subcellular localization and protein-protein interactions.

Additionally, we focus on applying new methods to study pathways and regulation at the cell type specific level. In collaboration with other lab members and external collaborators we perform single cell metabolomics and single cell transcriptomics to better understand how specialized metabolic pathways are organized at the cell type specific level.
An independent proof of concept study has focused on understanding better the protein level of regulators of cell type specific processes including but not limited to cell type specific pathway expression. Because detecting lowly abundant regulatory proteins is still a major challenge in single cell proteomics we use protein proximity labelling to pull down and analyze regulatory nuclear proteins from distinct cell types. This study has been performed in the model plant Arabidopsis thaliana because it is easily transformable and essential prior knowledge about cell type specific promoters is available.



