
Research
Plants produce an enormous number of complex molecules called natural products. These compounds play varied and rich roles in controlling the ways in which plants interact with the environment. Moreover, many of these compounds have life-saving medicinal properties and are used in both traditional and modern medicine. We are developing approaches to discover the genes and enzymes that are responsible for natural product biosynthesis, and we use this knowledge to understand the fundamental chemical, biological and evolutionary processes that underlie the biosynthesis of these complex molecules. Additionally, we develop platforms that allow fast and inexpensive production of these compounds.
Discovery of new plant biosynthetic genes
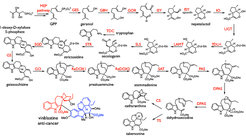
Many of the metabolic pathways that produce natural products in plants have not been characterized at the gene level.
We use state-of-the-art sequencing and bioinformatics, often in collaboration with the Buell group at MSU, along with chemical and biochemical approaches to rapidly identify new biosynthetic genes in plants.
To use our self organizing map developed by Marc Jones in Payne et al. (2017) Nature Plants:
Alkaloid biosynthesis
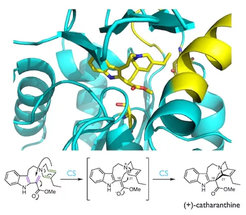
Alkaloids are nitrogen containing compounds that have varied and rich biological activities. The monoterpene indole alkaloids, are some of the most structurally complex molecules found in nature and include the pharmaceutically important compounds vincristine, quinine and strychnine.
Our group discovers and studies biosynthetic enzymes that create the chemical diversity found in the monoterpene indole alkaloids, which are found in five plant families.
For our sequence databases for some alkaloid producing plants:
Iridoid biosynthesis

Iridoids are a specialized class of monoterpenes best known for mediating interactions between plants and insects. Iridoids include the compounds known as nepetalactones, which are important not only for insect plant interactions, but are also the molecules in catnip that makes cats go wild.
Our group has discovered numerous enzymes of iridoid biosynthesis, and we are exploring how iridoid pathways evolved in plants such as catnip.
For our sequence databases for some iridoid producing plants:
Mechanism, evolution and function of biosynthetic pathways and enzymes
Many of the biochemical transformations that occur in natural product biosynthesis are poorly understood. Our group has discovered a number of unprecedented enzymatic transformations from plant metabolism, which we then study using mechanistic enzymology and structural biology approaches. In collaboration with Tours, we also look at protein-protein interactions in metabolism.
We also investigate how pathways may have evolved, using both genomic sequencing, as well as approaches such as ancestral sequence reconstruction.
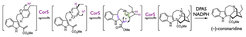
Metabolic engineering/synthetic biology of plant pathways
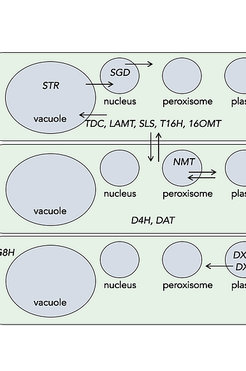
We create transgenic plant tissue capable of producing “new-to-nature” products. This reprogramming of biosynthetic pathways to produce unnatural products may lead to the development of molecules with new biological activities.
We are exploring ways to efficiently reconstitute individual branches of plant pathways in tractable host organisms, such as Nicotiana benthamiana and yeast, to improve production




