PhD Workshop of the Natural Products Section of the German Society for Plant Sciences
Date: Tuesday, March 29, 1:00-7:00 p.m., online via ZOOM
Participation is free and open to anyone interested, but please register first (see registration below)!
The program will consist of invited speakers and talks of doctoral researchers. If you are a doctoral researcher interested in contributing, please send a short (no more than one page) abstract to Jonathan Gershenzon (gershenzon@ice.mpg.de) or Angela Schneider (aschneider@ice.mpg.de) by February 20, 2022. Six of these will be chosen for talks during the meeting.
Invited Speakers
Schedule
13:00
Welcome - Ute Wittstock, Jonathan Gershenzon
13:05
Sarah O’Connor (Jena)
"How plants make complex alkaloids”
14:00
Paul Boemeke (Marburg)
“Identification and biochemical investigation of BAHD hydroxycinnamoyltransferases from Sarcandra glabra”
14:20
Simón Miranda (Trento)
“Integrating transcriptomic and metabolomic analysis to identify candidate genes involved in the biosynthesis of dihydrochalcones in Malus x domestica and wild Malus species”
14:40
Ling Chuang (Hannover)
“Elucidating quassinoid biosynthesis in the invasive Tree of Heaven reveals evolutionary origin from protolimonoids”
15:00
Jing-Ke Weng (Cambridge, MA)
“Harnessing plant metabolic evolution for new catalysts, medicines and materials”
16:00
Break
16:30
Andrea Müller (Jena)
“The formation of aldoximes and derivatives in a neotropical ant-plant”
16:50
Annika Engelhardt (Kiel)
“New insights into the biosynthesis of necic acids of lycopsamine type pyrrolizidine alkaloids”
17:10
Johann Hornbacher (Hannover)
“Indole glucosinolates are a source of auxin in drought-stressed Arabidopsis thaliana plants
17:30
Joe Jez (St. Louis)
“The sweet and sour of plant natural product biosynthesis”
18:45
Closing
Registration
Please register via ZOOM: https://zoom.us/webinar/register/WN_-3Rx8fJqR3ql6rjQFB_5eg

Sponsored by: Deutsche Botanische Gesellschaft, Sektion Pflanzliche Naturstoffe
Abstracts
Identification and biochemical investigation of BAHD hydroxycinnamoyltransferases from Sarcandra glabra (Thunb.) Nakai
Paul Boemeke, Maike Petersen
Institut für Pharmazeutische Biologie und Biotechnologie, Philipps-Universität Marburg, Robert-Koch-Straße 4, 35037 Marburg
As Sarcandra glabra (Thunb.) Nakai is one of a few species of the Chloranthaceae, an early angiosperm family, it is interesting for plant evolutionary research. Dry plant parts or their extracts are known in Traditional Chinese Medicine in form of teas or as medicinal products to treat joint pain or to reduce oxidative stress. The antioxidative, anti-inflammatory and antitumoral effects can be related to different natural compounds such as flavonoids or phenolic acids like chlorogenic acid and rosmarinic acid.[1] The biosynthetic pathway of rosmarinic acid is well known in Lamiaceae, like Coleus blumei (Cb), and Boraginaceae. [2] It is known that L-phenylalanine and L-tyrosine, which derive from the shikimate pathway, act as precursors. Both are modified differently: L-phenylalanine results in 4-coumaroyl-coenzyme A (pC-CoA) whereas L-tyrosine is converted to 4‑hydroxyphenyllactic acid (pHPL). A BAHD hydroxycinnamoyltransferase, rosmarinic acid synthase (RAS), transfers the hydroxycinnamic acid to the aliphatic OH group of pHPL.[3] Rosmarinic acid is the twice hydroxylated product of pC-pHPL.
Five different genes encoding CbRAS-like enzymes were found in S. glabra (SgHCT-A, -C, -D, -E, ‑F). The four full length nucleotide sequences SgHCT-A, -C, -D, -F were amplified from total RNA by polymerase chain reaction. After these sequences were introduced into Escherichia coli, the proteins were expressed and obtained from crude extract. The purified enzymes were used for biochemical assays. All assays were analysed via HPLC and additionally via LC-MS. In the first step, suitable substrates had to be found for each of the enzymes. The second step was to optimize reaction time, pH value and temperature. Km values were determined by modifying donor and acceptor substrate concentrations.
All four SgHCTs accept pC-CoA and caffeoyl-CoA (caf-CoA) as acyl donor substrates, whereas SgHCTs are varying in their acceptor substrate specificities. SgHCT-A can form hydroxycinnamic acid esters from shikimic acid, 3-hydroxyanthranilic acid and various hydroxy- or dihydroxybenzoic acids. However, SgHCT-C only shows activity with quinic acid. The precursor of rosmarinic acid, pC-pHPL, can be formed by SgHCT-D. A certain formation of caffeoylmethanol and caffeoylpropanol was detected in assays with SgHCT‑F containing methanol and 1-propanol, respectively.
The differences in substrate specificity indicate distinct functions of SgHCTs. All five nucleotide sequences can be found in total RNA which means the corresponding enzymes are actively involved in secondary metabolism. Yet, the level of activity has to be estimated by determining kinetic parameters. Besides a broad range of acceptor substrates, additional acyl donor substrates such as cinnamoyl-, feruloyl- and sinapoyl-CoA will be tested. As the proposed sequence of SgHCT-E lacks the 5´ end, RACE-PCR has to be done. Further investigation of rosmarinic acid metabolism in S. glabra needs to address the missing hydroxylation reactions. Responsible for hydroxylation of pC-pHPL to rosmarinic acid are cytochrome P450 enzymes of the CYP98A family. This knowledge will help to understand how pharmacological active phenolic acids in S. glabra are produced.
- Zeng Y, Liu J, Zhang Q, Qin X, Li Z, Sun G, Jin S (2021) The Traditional Uses, Phytochemistry and Pharmacology of Sarcandra glabra (Thunb.) Nakai, a Chinese Herb With Potential for Development: Review. Front Pharmacol 12:652926
- Petersen M (2013) Rosmarinic acid: new aspects. Phytochem Rev 12:207–227
- Petersen M (2016) Hydroxycinnamoyltransferases in plant metabolism. Phytochem Rev 15:699‑727
Integrating transcriptomic and metabolomic analysis to identify candidate genes involved in the biosynthesis of dihydrochalcones in Malus ´ domestica and wild Malus species
Simón Mirandaa, 2, Panagiotis Voulgarisa, Jorge Lagrèzea, Richard Espleyb, Andrew Darebb, Mickael Malnoya, Stefan Martensa
a Research and Innovation Centre, Fondazione Edmund Mach, Trento, Italy.
b The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand.
Dihydochalcones (DHCs) are specialized metabolites with a limited natural distribution, found in significant amounts in Malus x domestica (cultivated apple) and Malus wild species. Among them, M. x domestica accumulates significant amounts of phloridzin, whilst trilobatin and sieboldin are abundant in wild relatives. DHCs have demonstrated a wide range of bioactive properties in biomedical models. Some DHCs have also been reported to act as flavour sweeteners. Phloridzin may act as an anti-diabetic compound by blocking sodium-linked glucose transport and renal reabsorption of glucose in kidneys. Despite the protective effects reported in mammal models, little is known about how these metabolites are biosynthesised and what is their function in planta.
DHC pathway diverts from the main phenylpropanoid pathway from p-coumaroyl-CoA by the action of a postulated double bond reductase (DBR). Then, chalcone synthase (CHS) catalyses the condensation of p-dihydrocoumaroyl-CoA to phloretin. Phloretin can be directly glycosylated at position 2’- or 4’ to produce phloridzin or trilobatin, respectively. PGT1 and PGT2 have been identified as 2’- and 4’-O-UDP-glycosyltransferases responsible for the synthesis of phloridzin and trilobatin, respectively. However, sieboldin has been postulated to derive from hydroxylation in position 3 of phloretin before been glycosylated. In this study, we aim to identify candidate genes that may account for the reduction of the enoyl moiety (DBR) and the 3-hydroxylation of phloretin (3H), involved in DHC formation of apple and wild Malus species. Towards this aim, we combined transcriptomic and metabolomic analyses to identify candidate genes. Targeted metabolic profiling during leaf and fruit development of M. domestica and wild relatives (M. toringo, M. hybrid Evereste, M. micromalus) was conducted, and correlated with up-regulated DEGs of wild species following mapping to apple genome. Integrating omics data, we identified 24 putative DBRs and 2 potential 3Hs in each wild accession. We then assembled de novo transcriptomes of Malus species to retrieve the corresponding ORFs of putative DBRs and 3H. In addition, we stablished heterologous expression systems for de novo DHC production in E. coli, S. cerevisiae and model plants (Arabidopsis and N. tabacum) to functionally test candidate genes. We have cloned ORFs obtained in de novo transcriptomes, validating the assemblies obtained. Although assays in these systems are underway, preliminary results indicate that 2 potential DBRs were able to produce detectable amounts of phloretin in E. coli, one of which is currently being studied by CRISPR/Cas9 genome editing in apple. Altogether, these results provide an example of useful tools to prove the role of candidate genes involved in DHC biosynthesis in vivo.
Elucidating quassinoid biosynthesis in the invasive tree of heaven reveals evolutionary origin from protolimonoids
Ling Chuang, Shenyu Liu, Dave Biedermann and Jakob Franke
Leibniz University Hannover, Germany

The tree of heaven (Ailanthus altissima) is a globally invasive species that produces allelopathic compounds called quassinoids[1]. Quassinoids are highly modified triterpenoids with potent bioactivity for potential medicinal and agricultural application[2]. Even though being considered as taxonomic markers and defense metabolites of Simaroubaceae plants, how quassinoids are biosynthesized beyond mevalonate origin is completely unknown[3]. To explore the biological role of quassinoids in plants, elucidating their biosynthetic pathway is the first step to gain the basic knowledge for developing any biotechnological tool. Here, through sequence similarity network analysis of our de novo transcriptome data and heterologous expression in Nicotiana benthamiana, we identified and characterized the early steps of quassinoid biosynthesis in the tree of heaven. These steps show that quassinoids share a related origin with another group of highly oxidized triterpenes, limonoids, that are found in Rutaceae and Meliaceae from the same order Sapindales[4]. Our results not only show the first biochemical evidence of the protolimonoid origin for quassinoids but also provide the basis to further explore the biosynthesis and the biological roles of these complex triterpenoids in plants.
- M. Heisey, Am. J. Bot. 1996, 83, 192–200.
- I. J. Curcino Vieira, R. Braz-Filho, in Stud. Nat. Prod. Chem. , Elsevier, 2006, pp. 433–492.
- I. A. B. S. Alves, H. M. Miranda, L. A. L. Soares, K. P. Randau, Rev. Bras. Farmacogn. 2014, 24, 481–501.
- H. Hodgson, R. D. L. Peña, M. J. Stephenson, R. Thimmappa, J. L. Vincent, E. S. Sattely, A. Osbourn, Proc. Natl. Acad. Sci. U.S.A 2019, 116, 17096–1710
The formation of aldoximes and derivatives in a Neotropical ant-plant
Andrea T. Müllera, Yoko Nakamurab, Michael Reicheltc, Katrin Luckc, Eric Cosiod, Jonathan Gershenzonc, Axel Mithöfera and Tobias G. Köllnere
a Max Planck Institute for Chemical Ecology, Research group Plant Defense Physiology, D-07745 Jena, Germany
b Max Planck Institute for Chemical Ecology, Research group Biosynthesis/NMR, D-07745 Jena, Germany
c Max Planck Institute for Chemical Ecology, Department of Biochemistry, D-07745 Jena, Germany
d Pontifical Catholic University of Peru, Institute for Nature Earth and Energy (INTE-PUCP), San Miguel 15088, Lima, Peru
e Max Planck Institute for Chemical Ecology, Department of Natural Product Research, D-07745 Jena, Germany
Amino acid-derived aldoximes are precursors of many well-known specialized compounds such as cyanogenic glycosides, glucosinolates, and volatile nitriles and occur in many plant species. Additionally, aldoximes as such can provide direct defense against herbivorous caterpillars. Here, we studied the response to insect herbivory in the myrmecophytic plant Tococa quadrialata (Melastomataceae). Metabolomic analysis revealed a new aldoxime derivative accumulating in high amounts in the wounded leaves along with the corresponding free aldoxime. Transcriptomics enabled the identification and characterization of the enzymes involved in the biosynthesis of the aldoxime and its derived products in the ant-plant. A long-term study on the turn-over of the aldoxime and derivatives in the plant, together with findings on the metabolization of the derivative in the guts of insects in vitro, provide information about the function of the derivative. As we now found this aldoxime and its derivative accumulating in various plant species upon biotic stress, we speculate this might be a more general plant defense reaction.
New Insights into the Biosynthesis of Necic Acids of Lycopsamine Type Pyrrolizidine Alkaloids
Annika Engelhardta, Dorothee Langela, Sebastian Fritza, Britta Muhsa and Dietrich Obera
aBotanical Institute and Botanic Gardens, Christian-Albrechts-University, Am Botanischen Garten 5, 24118 Kiel, Germany
Pyrrolizidine alkaloids (PAs) are typical specialized plant metabolites involved in plant defense. The backbone of PAs consists of the characteristic bicyclic necine base that is esterified with one or more necic acids. The great structural diversity of the PAs with more than 600 different structures has its origin mainly in the diversity of the necic acids [1]. While the biosynthesis of the necine base moiety has been intensively studied over the past decades our knowledge about the biosynthesis of the necic acids is scanty. Lycopsamine type PAs have at least one necic acid with a C7- carbon skeleton that is unique within angiosperms[1]. Earlier feeding experiments with root cultures of Eupatorium clematideum (Asteraceae) using [13C6] glucose and 13C13C coupling analyses within the necic acid moiety suggested, that a C2- moiety is transferred from hydroxyethyl thiamine diphosphate to 2- oxoisovaleric acid (2- OIV) by an acetohydroxyacid synthase (AHAS, EC 2.2.1.6)- like enzyme to generate the C7- necic acid backbone[2]. AHAS catalyzes the transfer of a C2- moiety to pyruvate or 2- oxobutyrate early in the branched-chain amino acid biosynthesis. It is composed of catalytic subunits (CSUs) that are responsible for the biochemical reaction and regulatory subunits (RSUs) that are responsible for feedback inhibition [3].
Using sequence data of various AHAS and a reverse genetic approach we identified two CSUs and one RSU of AHAS in Symphytum officinale. The most likely candidate, SoCSU2, was determined by RT-qPCR. CRISPR/Cas9- mediated knock-outs provided in planta evidence that this candidate is indeed responsible for the formation of the necic acid backbone of lycopsamine type PAs. Subsequently, both CSUs were heterologously expressed in E. coli and compared with respect to their biochemical properties. For this purpose, we developed an assay for AHAS that allows the identification and quantification of substrates and products simultaneously. This assay was used to estimate kinetic parameters and substrate preferences of the two CSUs allowing us to distinguish between the enzyme involved in the biosynthesis of branched-chain amino acids and that involved in the biosynthesis of the characteristic C7- moiety of lycopsamine type PAs.
- Hartmann and Witte (1995)
- Weber et al. (1999)
- Duggelby and Pang (2000)
Indolic glucosinolates are a source of auxin in drought-stressed Arabidopsis thaliana plants
Johann Hornbacher, Ina Horst-Niessen, Jutta Papenbrock
Institute of Botany, Leibniz University Hannover, Herrenhäuser Str. 2, D-30419 Hannover, Germany
Members of the Brassicales are capable to synthesize indole-3-acetic acid (IAA) from the indolic glucosinolate (iGSL) glucobrassicin (GB) (Fig.). To investigate the role of this particular pathway during abiotic stress, Arabidopsis Col-0 and several mutants lacking key enzymes in this pathway were subjected to drought stress (DS). Indolic metabolites and the expression of genes involved in this pathway were analyzed. To investigate the synthesis rate of GSLs, plants were subjected to deuterium oxide (D2O) in an additional experiment and incorporation of deuterium into GSLs was monitored using LC MS. Additionally, fitness of plants was assessed by analyzing contents of reactive oxygen species (ROS).
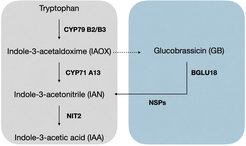
enzymes.
Contents of GB were significantly higher in shoots of DS compared to control plants. Higher incorporation of deuterium into GB revealed a higher turnover compared to other GSLs. Elevated expression of the thioglucosidase BGLU18 in DS plants suggests its involvement in the breakdown of GB (Fig.). Significantly higher contents of ROS were observed in DS bglu18 mutants compared to controls highlighting its importance in plant fitness. Furthermore, higher contents of GB were observed in DS bglu18 mutants compared to DS Col-0 indicating the compensation of the loss of BGLU18 with elevated levels of GB. Expression of CYP71A13, encoding for a key enzyme in the synthesis of IAA independently of GB, was observed to be higher in DS bglu18 compared to controls. This indicates the compensation of the compromised GB pathway by using other synthesis pathways yielding IAA in bglu18. Higher expression of NSPs encoding for nitrile specifier proteins and higher contents of indole-3-acetonitrile (IAN) in DS compared to control plants highlight the direction of the degradation towards the formation of IAN. Significantly higher contents of ROS were observed in DS nsp1 mutants compared to controls and Col-0 underlining the compromised fitness in control and DS conditions. Finally, higher expression of NIT2, encoding for a nitrilase, and elevated contents of IAA in DS plants compared to control plants indicate the formation of IAA from GB in DS plants.











