Neurogenetics
Binary systems of expression
To express a given gene of interest in a specific cell/tissue pattern, we make use of complimentary binary systems of expression - GAL4-UAS (upper left graph) and lexA-lexAop (upper right graph) - that encode transactivators and responder elements, separately. A driver fly-line expresses the yeast transcription activator protein GAL4 (upper left graph), or a chimeric transcription activator protein formed by the bacterial repressor LexA fused to the C-terminal activation domain of GAL4 (upper right graph), under the regulation of a specific promoter/enhancer. A responder fly-line carries a gene of interest (gene X) under UAS (Upstream Activation Sequence, upper left graph), an enhancer to which GAL4 specifically binds to activate gene transcription, or under a gene-specific LexA DNA-binding motifs (LexAop, upper right graph). When a driver line is crossed to a corresponding responder line, the progeny expresses the gene of interest (gene X) under the regulation of the specific promoter/enhancer. A third type of driver fly line (lower graph) expressing GAL80ts, a yeast temperature-sensitive GAL4 repressor, provides temporal resolution to the GAL4-UAS system. Under the restrictive temperature (18°C) GAL80 binds to the transcriptional activation domain of GAL4 blocking its activity; when flies are transferred to the permissive temperature (29°C), GAL80 frees GAL4 to activate gene expression acting via UAS.

Transcriptomics / RNAseq
Gene families involved in the detection of odorants exhibit tremendous sequence variability, and large gene numbers in most species. Our lab has started the use of transcriptomic sequencing several years ago to allow the identification and study of evolution of the respective gene families, especially in species which are relevant from the viewpoint of evolutionary biology, but are not model species with existent datatsets.

Deorphanization of receptors based on expression alternations of mRNA levels (DREAM)
One important first step for many neurogenetic approaches in olfaction is the association of odorants and detecting receptors. Published by von der Weid et al., in 2015, the DREAM method utilizes detection of receptor mRNA regulation after prolonged odor stimulation to this end. We have established use of DREAM, detecting receptor coding transcripts with qPCR, RNAseq or Nanostring technology. A key feature of the method is the focus on the applied odor – we do not ask the question ‚what does this receptor do?‘, but ‚which receptors are important for the detection of this behaviourally/ecologically relevant odor?‘.

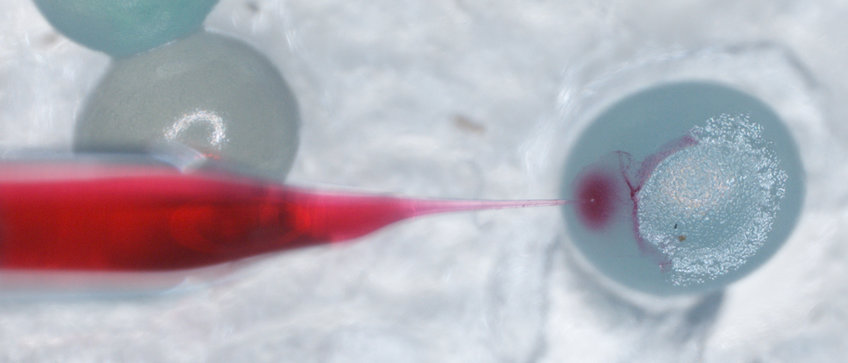
CRISPR Cas9
Revolutuionizing the field of gene manipulation, targeted gene editing in vivo using CRISPR Cas9 has been making headlines in recent years. We have established the method for use in the tobacco hornworm Manduca sexta, allowing us to connect genes, gene function and behaviours.




