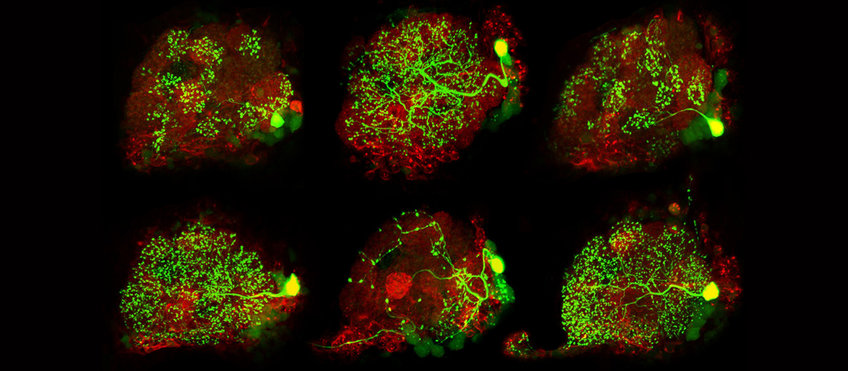
Neuroanatomy
Photoactivation
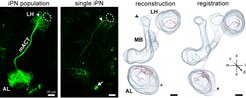
In order to analyze neurons at the single cell level, we perform neural tracing by employing a genetically encoded photoactivatable GFP (PA-GFP). PA-GFP is photoconverted with a continiuous illumination of 760 nm which results in a 100-fold increase of its GFP fluorescence. By photoactivating single somata with an IR laser at the 2-photon microscope, individual neurons can be selectively labeled from their soma up to their farthest axonal terminals. Afterwards, a z-stack of the whole brain will be acquired and subsequently used for neuronal 3D-reconstructions. All reconstructed neurons will be transformed and registered into our standard in vivo brain to align neurons of different individuals.
Strutz et al. eLife. 2014
Grabe et al. CellReports. 2016
Neuronal Tracing
We are employing computer-assisted, semi-automated digital techniques to map neuronal features and circuits in the brain. Image data are acquired by confocal laser scanning (CSLM), electron microscopy (TEM, FIB-SEM) as well as neuronal tracings correlated with physiological experiments (optical imaging, electrophysiology, and patch-clamp). Using the 3D reconstruct software Amira-Software we are semi-automatically tracing the neuron morphology and segment the corresponding brain regions (neuropils). Quantitative morphometry allows us for statistical analysis of common neuron parameters. In combination with Image Registration techniques morphological data can be linked to physiological and molecular data and compared across species.
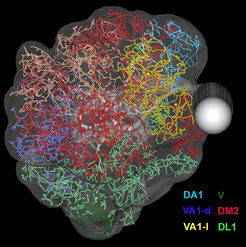
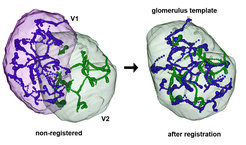
Image Registration
Image Registration is a commonly used technique in order to visualize neuronal morphologies in a common 3D reference space. In this coordinate transformation process coordinates of one image are mapped onto the anatomically equivalent point in another image. We employ grey-value-, labelfield, and landmark-based registration using the software Amira (FEI?). Eventually, registration allows us to analyze and compare data across different modalities and experiments in a common reference system (Rybak, 2016).
GRASP
GRASP (GFP reconstitution across synaptic partners) is a genetic technique that allows the identification of synaptic partners in genetically amenable animals by labeling putative synaptic contacts between two or more neurons in the brain. The GRASP method is based on a protein complementation of two fragments of green fluorescent protein (GFP) fused to extracellular domains of transmembrane proteins.
We are using synaptic-tagged GRASP constructs in which spGFP1-10 is fused to Neurexin (a transmembrane protein specific for synaptic sites) and spGFP11 fused to the membrane. We make use of two different binary transcriptional systems: Gal4-UAS and LexA-LexAop to express each GFP fraction in two different neuronal populations.
The GRASP signal is visualized in vitro after immunolabeling with light microscopic techniques.

Electron Microscopy
Scanning Electron Microscopy (SEM) is employed in order to visualize and quantify cuticle structures, such as sensory sensilla, in different insect species (Manduca sexta, Drosophila melanogaster, and Crustacea). The technique allows depicting external morphological features at a high resolution.
Classical Transmission Electron Microscopy (TEM) is employed in correlation with confocal microscopy of genetically labelled neurons to study the synaptic connectivity in Drosophila melanogaster. Ultrathin sections are cut manually, collected on grids and then photographed at the TEM. Serial, consecutive sections are aligned and neuronal profiles are segmented using the open source software Trak-EM (Image J-Fiji). 3D rendering techniques allow to collectively visualizing synaptic networks in 3D-PDF format. (Rybak et al., 2016).
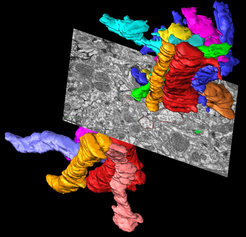
In addition, we are using large-scale automated EM techniques, such as Focused ion Beam-Scanning Electron Microscopy (FIB-SEM) in order to gain complete and fast reconstructions of neuronal networks at the ultrastructural level. Laser branding (using two-photon laser scanning microscopy) is used in order to identify the brain region of interest at the ultrastructural level. For data collection open-source software such as CatMaid http://openconnecto.me/catmaid/ are used.

LightSheet Microscopy

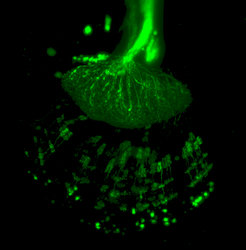
Light sheet fluorescence microscopy (LSFM) is a 3D tomographic technique in which a thin light sheet is used in contrast to the point laser scanning of confocal microscopy. Advantages are fast scanning times and high contrast that allows high-resolution scans in space and time. The fast scanning reduces photo damage and is therefore most suited for in-vivo imaging of fluorescent labelled structures.
Currently we are employing this technique in order to gain fast scans of the whole brain and of the external cuticle in diverse insect species.







